Past Pilot Programs
The fate and function in capillary development of the peri-ureteric bud pericytes in the developing mouse renal medulla
Pilot Project 1: The fate and function in capillary development of the peri-ureteric bud pericytes in the developing mouse renal medulla
Principal Investigator: Jing Yu, Ph.D.
Mentor: Ariel R. Gomez, M.D.
The capillary network of the mammalian renal medulla plays a key role in the regulation of body water and electrolyte homeostasis which is critical for extrarenal wellness such as cardiovascular health and normal brain function. Despite its physiological importance, our knowledge on where this capillary network arises from and how they form is largely unexplored. Understanding the origin and the mechanisms underlying the formation of this vasculature network is not only important for the establishment of a functional kidney at the beginning of postnatal life, but will also facilitate efforts in repair and rebuilding of a renal medulla following damage and disease both in vivo and in vitro. Our previous work revealed that Wnt7b signaling from the ureteric bud (UB) epithelium regulates lumen formation of the peri-UB capillaries in the nascent renal medulla of the developing mouse kidney. Wnt7b signals to the pericytes associated with this population of the capillaries, suggesting that these pericytes play a role in lumen formation of the capillaries that they are associated with. Our long term objective is to understand the fate of this population of pericytes in the adult stage in normal and abnormal conditions and the role they play in regulating capillary lumen formation. In this application, we propose to test the hypothesis that the pericytes of peri-UB capillaries in the developing renal medulla contributes to pericytes associated with the vasa recta in the adult stage and they act on endothelial cells to regulate the lumen formation of the medullary peri-UB capillaries (Figure 1). We will test this hypothesis in the following two specific aims,
SA1. Determine the fate of the medullary peri-UB capillary pericytes at the adult stage
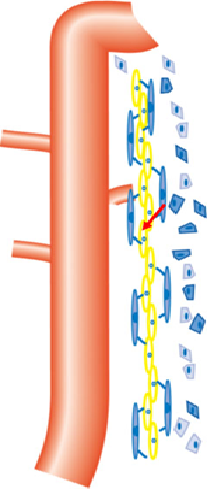
Fig. 1. A schematic diagram of the medullary peri-UB capillaries. The orange tube is the UB epithelium. The long blue cells are pericytes, and the long yellow cells are endothelial cells of the peri-UB capillaries. The blue polygon cells are interstitial cells.
In the developing mouse kidney, Neurotensin (Nts) is expressed in the renal medullary interstitium surrounding the ureteric bud epithelium, the pericytes of medullary peri-UB capillaries. A Nts-Cre mouse strain where the Cre recombinase is expressed from the endogenous Nts locus is available for us. We will employ Nts-Cre; mT/mG mice in which the Cre expressing cells and their progeny express GFP at the cell membrane. We will determine whether the GFP-expressing cells in the kidney at the adult stage are pericytes associated with vasa recta with co-immunostaining for GFP and vasculature markers such as Pdgfrb and Desmin (pericyte) and UT-B (descending vasa recta).
SA2. Identify signals from the pericytes to endothelial cells that promotes capillary lumen formation.
We will sort out GFP+ cells from E15.5 Nts-Cre; mT/mG kidneys with Fluorescence-activated cell sorting (FACS), and perform single cell RNA-seq to identify, bioinformatically, genes encoding secreted proteins in these cells, especially in those cells that express Nts and p57Kip2 (pericytes of medullary peri-UB capillaries). We will then perform in situhybridization or immunofluorescence staining to validate the expression in the pericytes of medullary peri-UB capillaries of these genes, with the top priority for genes implicated in vascular development in the literature.
We will also develop a 3-D endothelial cell-pericyte co-culture system with HUVEC or endothelial colony forming cells (ECFC)-derived endothelial cells and the GFP+ cells sorted from E15.5 Nts-Cre; mT/mG kidneys.
Lastly, we will screen the secreted factors, in particular genes implicated in vascular development in the literature, identified with RNA-seq and validated experimentally for pericytes derived signals that promote endothelial lumen formation, using the 3-D co-culture system. We will determine if endothelial tube lumen formation is disrupted with blocking antibodies or small molecule inhibitors for these gene products, where available. Alternatively, if the secreted proteins are
available, we will apply the proteins to the 3-D HUVEC or ECFC-derived endothelial cell culture to see if they promote endothelial tube lumen formation.
The proposed work will serve as the basis for determining the fate of these pericytes and their progenies in renal diseases (acute and chronic kidney injuries) and repair, genetic analysis to determine the role of pericytes in medullary peri-UB capillary lumen formation in vivo, and for engineering renal medullary capillaries in vitro.
The role of Pappa2 in kidney development
Pilot Project 2: The role of Pappa2 in kidney development
Principal Investigator: Vikash Kumar, PhD
Mentor: Allen W. Cowley Jr, PhD
Hypothesis and Specific Aims
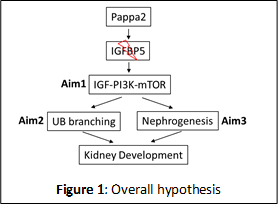
The goal of this grant is to study the function of Pappa2 in the development of kidney and in particular ureteric bud (UB) branching and nephrogenesis in rats and mice. Pappa2 (pregnancy associated plasma protein A2) is a protease of insulin-like growth factor binding protein 5 (IGFBP5) which has been primarily studied in pregnancy and postnatal growth. Our recent findings in salt-sensitive hypertension found that a subcongenic Dahl salt-sensitive (SS) rat strain containing a 0.71 Mbp of chromosome 13 which includes Pappa2 gene from salt-insensitive Brown Norway (26-P strain) is protected significantly (24 mmHg) from salt-induced hypertension. Contained within this narrow congenic region resides three genes Pappa2, Astn1 and miR-488. Only Pappa2 was found to be differentially expressed in the renal cortex and altered by salt diet intake. This observation focused our attention on to investigate the potential role of Pappa2 in the kidney about which nothing is known.
Pappa2 gene encodes a member of the pappalysin family of metzincin metalloproteinase which cleaves IGFBP5 and regulates insulin-like growth factor (IGF) bioavailability and downstream signaling. It is recognized that Pappa2 modulates development of bone size, cranial cartilage and angiogenesis. Recently, homozygous loss‐of‐function mutations in Pappa2 were reported in human subjects which resulted in a novel syndrome of growth retardation with markedly decreased free IGF‐I concentrations and IGF bioactivity. There is evidence that Pappa2 is expressed in the kidneys of mice and rats. While the functions of IGFBP-5 and its ligands, the IGFs, have been well-studied, the in vivoregulation of the IGFBP5 by Pappa2 is poorly understood and the functional significance of these events in the kidney is unknown. We hypothesized that Pappa2 determines UB branching and nephrogenesis by regulating the IGF/PI3K/mTOR signaling in the kidney development. Preliminary data supporting our hypothesis:
- Pappa2 found to be regulate IGF/PI3K/mTOR signaling in vitro.
- Pappa2 is differentially expressed in the developing kidney of SS and 26-P congenic rats.
- It is localized in the ureteric bud (UB) and cortical thick ascending limb (cTAL).
- 26-P congenic rats exhibited greater glomerular number compared to age-matched SS rats.
- CRISPR/Cas9 knockout of the Pappa2 gene in 26-P congenic rats exhibited a significant reduction in body weight and glomerular number compared to littermate wildtype rats.
- Conditional KO of Pappa2 in the nephron progenitors of mice has no significant effects on body weight and glomerular number.
The above hypothesis (Figure 1) will be examined in three specific aims:
Specific Aim 1: Determine the role of Pappa2 in the regulation of IGF/PI3K/mTOR pathways in vivo by studying the Pappa2 KO rats and littermate WT control.
Specific Aim 2: Determine the effect of Pappa2 on UB branching by examining ex vivo kidney culture of Pappa2 KO and WT rats in the presence or absence of Pappa2, IGFBP5 and mTOR inhibitors.
Specific Aim 3: Determine the function of Pappa2 in nephrogenesis by investigating the IGF/PI3K/mTOR pathways, cell proliferation and apoptosis in conditional KO of Pappa2 mice and littermate WT mice.
Rapid identification of master regulators of renin cell fate: from fish to mice
Pilot Project 3: .
Principal Investigator: Bernard V. Thisse, Ph.D. and Christine I. Thisse Ph.
Statement of the scientific problem and preliminary results
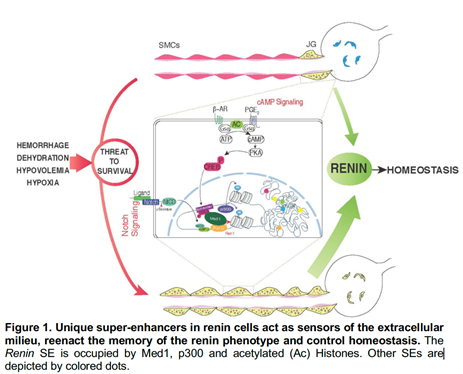
Renin-expressing cells are crucial for survival: they control fluid-electrolyte and blood pressure homeostasis, vascular development, glomerular regeneration, and oxygen delivery to tissues. In mice, during embryonic development, renin cells are progenitors for arteriolar smooth muscle, and glomerular mesangial cells. When there is a threat to survival, those descendants (which retain the memory of the renin phenotype) undergo a remarkable transformation from contractile to endocrine renin-synthesizing cells until homeostasis is regained (Figure 1). The Gomez lab recently found that the identity of renin cells may be determined by unique super-enhancers (SEs), dynamic regions of chromatin that are associated with lineage- and cell-specific genes (Figure 1).
Structurally, SEs are characterized by their relaxed-accessible chromatin, deposition of histone marks such acetylation of lysine 27 of Histone 3 (H3K27ac) characteristic of active enhancers, heavy deposition of transcription factors (TFs), binding of the mediator complex, and the acetyl transferase p300. The top ranked SE shared among all mammalian renin cells examined was found at the Renin locus. This SE controls Renin expression and the molecular memory of the renin phenotype, a fundamental process to ensure survival when homeostasis is threatened.
In the pursuit of this work, the Gomez lab also identified a core set of 171 transcription factors (out of »2000) that may be crucial for the activation of the SEs and the memory of the renin phenotype. Assessing the functional relevance of such a large number of TFs in mice, in vivo, will be impractical, costly and extremely slow. Fortunately, the functions and regulation of renin are highly conserved between fish and mammals and it is likely that master regulators of renin cell identity and function are equally conserved between the two species.
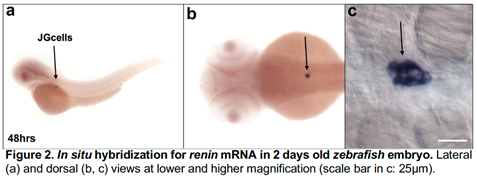
Figure 2 shows the exquisite localization of renin mRNA in the JG cells of a zebrafish embryo at 48 h post fertilization. Because zebrafish embryos are transparent they also allow the visualization of fluorescent proteins driven by the renin regulatory regions. Due to their rapid development, transparency and conservation of function combined with the ability to generate large number of fish embryos, it is now possible to rapidly screen the functional relevance of a large number of putative master transcription factors with the highest likelihood of determining the fate of these cells in both species.
Hypotheses, scientific approach, pitfalls and alternatives
In collaboration and under the mentoring of Dr. Maria Luisa Sequeira Lopez and Dr. Ariel Gomez (Child Health Research Center, UVA), experts in renin cell biology we will use the methodologies uniquely available in our respective laboratories and propose to test the following hypotheses:
- Mice and fish share a core set of ancestral SEs that characterize the fate of JG cells in both species. JGs will be isolated from embryonic and adult fish harboring a renin promoter linked to Red Fluorescent Protein (RFP). The distribution of RFP labelled cells is similar to the distribution of renin mRNA during development and in response to physiological challenges indicating that RFP labels bona fide JG cells and that the regulation of renin is similar to the one observed in mammals. Cells will be sorted as previously described in Gomez lab and processed for single cell RNA-seq (sc-RNA-seq) using our Fluidigm C1 machine located at the CHRC. To determine whether the fish cells share the same SEs as the mammalian JG cells a group of cells will be processed for Chip-seq for H3K27ac, ATAC-seq (to detect areas of chromatin open for transcription) and p300 deposition. The results will be compared to those already available in Gomez lab mouse JG cell database. We expect that fish and mouse cells will share some ancestral SEs including the characteristic renin SE. Those SEs will in turn be the site for binding of some crucial TFs. Comparison of TFs found in the mouse JG cells with those found in the zebrafish JG cells will allow us to identify those that are more likely to play a role in renin cell specification. The expression pattern of those TFs in the zebrafish will be assessed by in situ hybridization and those specifically labeling JG cells will be chosen for functional screening as described below.
- A core group of TFs are functionally responsible for the specification of the JG cells. To test this hypothesis, the TFs identified in Aim 1 will be knocked down either one by one or in combination using splice-interfering morpholino that allow for the evaluation of knockdown efficiency using RT-PCR and following the guidelines for morpholino use in zebrafish. Morphant embryo will be analyzed by in situ hybridization at 2 days of development when the JG cells are the only cell expressing renin in the embryo (Figure 2). It is expected that ablation of the TFs will result in disappearance of the renin in situ hybridization signal. However, if this would not be the case, morpholino injections will be performed in Tg(ren:RFP) embryos and the expression of RFP under the renin promoter will be monitored and quantified based on the intensity of fluorescence at 2 days of development and compared to the intensity of fluorescence of uninjected embryos at the same stage of development. After this initial screening, we will inhibit the action of those TFs that seem to be functionally relevant using a CRISPRi approach to block the action to those TFs without the need to delete or mutate the underlying DNA. Alternatively, by multiplexing sgRNAs into a single injection performed in embryos of the Tg(ren:RFP) line we will generate biallelic mutations that will be analyzed by looking at the expression of the RFP, observed either under fluorescence or by in situ Positive results observed in F0 embryos will be confirmed by generating the mutant line and analyzing the expression of renin in homozygous mutant embryos.
Once the search is narrowed to a few functionally relevant TFs in zebrafish, the same TFs will be deleted in mice using a cre-lox approach as previously described for Ren1d-cre mouse line.
Relevance and application of the proposed work
By uncovering ancestral, conserved mechanisms that determine cell identity, the proposed work will be applicable not only to how cell fate is regulated in the renin cell, but it will generate fundamental information applicable to similar work in other cell types such as the insulin secreting cell, the erythropoietin producing cell and other cell types in the body. The results of these studies will also generate important preliminary data for extramural grant (RO1 and PO) applications to NIDDK and NHLBI. We expect the results of these studies will benefit children and adults with renal and cardiovascular diseases and improve our efforts in Regenerative Medicine.